Forschungsprofil PD Dr. Oliver Bader
- Leitung AG Mykologie
- IT-Koordination des Instituts
- Mikrobiologisch diagnostisches Massenspektrometrielabor
- Kurator der internationale MLST-Datenbank für Candida glabrata (pubmlst.org/).
- Stellvertretender Ringversuchleiter Candida-Serologie (Instand e.V.)
Kontakt: +49 551 39 65403; obader(at)gwdg.de
Mein Team und ich beschäftigen uns hauptsächlich mit verschiedenen Aspekten der Epidemiologie, Diagnostik und Pathogenese klinisch relevanter Pilzinfektionen des Menschen [vollständige Publikationsliste], arbeiten aber auch in Abhängigkeit der Fragestellung mit verschiedenen bakteriellen Erregern.
Je nach Kapazität sind wir für Laborrotationen sowie Bachelor- und Masterarbeiten aus den molekularbiologisch orientierten Studiengängen der Universität Göttingen offen, ebenso bieten wir auch Plätze mit hohem praktischem Laboranteil für medizinische Promotionen an.
Arbeiten zu Aspergillus fumigatus
Bei dem humanpathogenen Schimmelpilz Aspergillus fumigatus stellen erworbene Resistenzen gegenüber klinisch genutzten Azolwirkstoffen weltweit ein zunehmendes klinisches Problem bei schwerkranken Patienten dar. Ein Großteil der Resistenzen ist auf reduzierte Affinität des Zielenzyms Cyp51A gegenüber Azolwirkstoffen zurückzuführen. Neben den klassischen cyp51A-Mutationen an den Positionen G54 und M220, welche auch durch Langzeittherapie entstehen können, haben retrospektive Analysen gezeigt das den frühen 2000er Jahren auch pandemische Allele auftreten, welche sich in der Umwelt verbreiten. Diese weisen neben Mutationen in der proteinkodierenden Sequenz auch Veränderungen im Promotorbereich auf, welche die erhöhte Expression von cyp51A zur Folge haben. Für solche Veränderungen wurde postuliert, dass sie durch Einwirken landwirtschaftlich genutzter Azol-basierter Fungizide induziert wurden.
Bereits in frühen Studien für den deutschsprachigen Raum konnten wir zeigen, dass diese Stämme sowohl in der Umwelt als auch Patienten auftreten, und auch erstmalig das Auftreten der verschiedenen sich verbreitenden Promotorvarianten (TR34, TR46) in Deutschland nachweisen. Mittlerweile zeigen weltweit viele weitere Studien, zu denen auch wir in Kooperationen mit Partnern in Thailand und Ostafrika beigetragen haben, dass sich diese Resistenzallele global verbreiten. In unseren Arbeiten wurde unerwarteter Weise auch deutlich, dass sich auch Allele mit Veränderungen an G54 und M220 in der Umwelt finden lassen. Ob solche resistenten Stämme aus dem Krankenhausbereich stammen und wie sie in der Umwelt vermehrt werden können ist derzeit noch unklar.
Aktuelle Arbeiten befassen sich neben der Epidemiologie auch immer wieder mit der Diagnostik solcher Resistenzen. In der Vergangenheit waren dies v.a. die Entwicklung von Resistenz- und PCR-Nachweisen spezifischer Mutationen. Im Hinblick auf den Einzug neuer Hochdurchsatz-Sequenzierungstechnologien in die mikrobiologische Routinediagnostik widmen wir uns im Moment verstärkt der systematischen Auswertung von Mutationen (SNPs, Allelvarianten) in Resistenzgenen (Kooperation Dr. Christoph Sasse), um diese für die Sequenzanalyse bewertbar zu machen.
Ausgewählte Publikationen:
Bader O (Nov 2021) Phylogenetic Distribution of csp1 Types in Aspergillus fumigatus and Their Correlates to Azole Antifungal Drug Resistance. Microbiol Spectr. 17:e0121421. doi: 10.1128/Spectrum.01214-21. [Pubmed]
EA Idelevich EA, Groß U, Becker K, und Bader O (May 2018). Comparative evaluation of different gradient diffusion tests for detection of azole resistance in Aspergillus fumigatus. Diagn Microbiol Infect Dis 91(1):52-54. doi:10.1016/j.diagmicrobio.2018.01.003. [Pubmed]
Dudakova A, Spiess B, Tangwattanachuleeporn M, Sasse C, Buchheidt D, Weig M, Groß U, und Bader O (Oct 2017). Molecular tools for the detection and deduction of Aspergillus azole resistance phenotypes. Clinical Microbiology Reviews 50(4):1065-1091. doi: 10.1128/CMR.00095-16. [Pubmed]
Tangwattanachuleeporn M, Minarin N, Saichan S, Sermsri P, Mitkornburee R, Groß U, Chindamporn A, and Bader O (Jun 2017). Prevalence of azole resistant Aspergillus fumigatus in the environment of Thailand. Med Mycol. 55(4):429-435. doi: 10.1093/mmy/myw090. [Pubmed]
Bader O, Tünnermann J, Dudakova A, Tangwattanachuleeporn M, Weig, und Groß U(2015). Environmental isolates of azole-resistant Aspergillus fumigatus in Germany. Antimicrob Agents Chemother 59(7):4356-9. [Pubmed]
Arbeiten zu Candida albicans
Candida albicans ist ein kommensaler Hefepilz im Gastrointestinaltrakt vieler Säugetiere, darunter auch dem Menschen. Bei gesunden Menschen besteht ein gut ausbalanciertes Gleichgewicht zwischen dem Immunsystem, dem Pilz und den anderen Mikroorganismen des Mikrobioms. Kommt dieses Gleichgewicht aus der Balance, etwa durch antibakterielle Wirkstoffe oder Störungen im Immunsystem wie bspw. einer Chemotherapie, kann der Pilz sein pathogenes Potential entfalten und schwerwiegende, systemische Infektionen verursachen. Candida albicans zeichnet sich wie auch dadurch aus, dass er in vielen verschiedenen Zellmorphologien wachsen kann, die alle eine spezifische Rolle in den verschiedenen zu besiedelnden Nischen im Wirt spielen. Alle Formen haben stark unterschiedliche zelluläre Anforderungen, und daher auch unterschiedliche Transkriptom- und Proteom-Profile.

In laufenden Arbeiten versuchen wir zu verstehen, wie C. albicans es schafft, das mit der Ausprägung von Virulenzeigenschaften assoziierte Myzelwachstum während der kommensalen Phase zu unterdrücken und dies möglicherweise auf epigenetischer Ebene zu verankern
Ausgewählte Publikationen:
Mühlhausen S, Schmitt HD, Plessmann U, Sternisek P, Perl T, Weig M, Urlaub H, Bader O, und Kollmar M (Dec 2021) The Candida albicans CUG codon is translated as unambiguously as every other codon. BMC Biol 19, 258. doi: 10.1186/s12915-021-01197-9. [Pubmed]
Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Förster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Häder A, Kurzai O, Luo T, Krueger K, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, und Naglik JR (2016). Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532(7597):64-8. Doi: doi: 10.1038/nature17625. [Pubmed]
Arbeiten zu anderen Candida-Hefen
Neben Candida albicans kommen auch andere Candida-Spezies als Besiedler und Pathogen beim Menschen vor, darunter auch Candida parapsilosis und die phylogenetisch etwas weiter entfernte Hefe Candida glabrata. Bei beiden Hefen interessiert uns vor allem der Aufbau der Zellwand (Kooperation mit Prof. M. Weig und Prof. PW deGroot) und die daraus resultierende Eigenschaft Biofilme zu bilden. Candida glabrata zeichnet sich durch eine Vielzahl unterschiedlicher potentieller Adhäsin-Gene aus, die in der Population unterschiedlich verteilt sind.
Durch Zuordnung unterschiedlicher phänotypischer Eigenschaften auf Populationsebene versuchen wir daraus potentielle Gewinne für Diagnostik und Therapie durch Subtypisierungen abzuleiten. In diesem Zusammenhang kuratiere ich seit 2016 auch die internationale MLST-Datenbank für Candida glabrata (pubmlst.org/).
Ausgewählte Publikationen:
B Zhai, C Liao, S Jaggavarapu, Y Tang, T Rolling, Y Ning, T Sun, SA Bergin, M Gjonbalaj, E Miranda, NE Babady, O Bader, Y Taur, G Butler, L Zhang, JB Xavier, DS Weiss, and TM. Hohl (2024) Echinocandin heteroresistance causes prophylaxis failure and facilitates breakthrough Candida parapsilosis infections. Nat Medicine (2024). doi: 10.1038/s41591-024-03183-4 [Pubmed]
KP Wojak, G Ungermann, I Ichsan, E Gomez-Molero, K Jung, M Weig, F Nauck, D Ziebolz, Y Gräser, U Groß, B Alt-Epping, and O Bader (Jul 2021) Host age and denture wearing jointly contribute to oral colonization with intrinsically azole-resistant yeasts in the elderly. Microorganisms 9(8):1627. doi: 10.3390/microorganisms9081627. [Pubmed]
E Gómez-Molero, I De-la-Pinta, U Groß, M Weig, G Quindos, P deGroot§, and O Bader§ (Jan 2021) Candida parapsilosis colony morphotype forecasts biofilm formation of clinical isolates. J Fungi (Basel) 7(1):33. doi:10.3390/jof7010033. [Pubmed]
T Gabaldón, E Gómez-Molero, and O Bader (Oct 2020) Molecular typing of Candida glabrata. Mycopathologia 185(5), 755-764. doi: 10.1007/s11046-019-00388-x. [Pubmed]
E Gomez-Molero, JR Willis, A Dudakova, A Gacser, M Weig, U Groß, T Gabaldón, and O Bader (Aug 2020) Phenotypic variability in a coinfection with three independent C. parapsilosis lineages. Front Microbiol 11:1994. doi:10.3389/fmicb.2020.01994. [Pubmed]
L Carreté, E Ksiezopolska, E Gómez-Molero, A Angoulvant, O Bader, C Fairhead, and T Gabaldón (Feb 2019) Genome comparisons of Candida glabrata serial clinical isolates reveal patterns of genetic variation in infecting clonal populations. Front in Microbiol. 10:112 doi:10.3389/fmicb.2019.00112. [Pubmed]
§: „equal contribution“
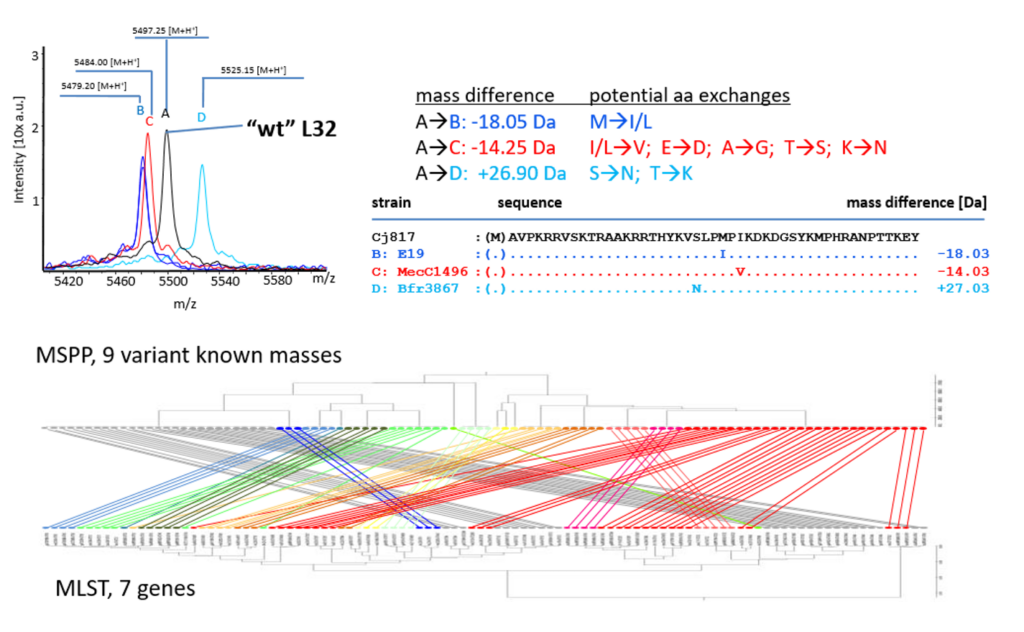
MALDI-TOF MS & IT
In meinen weiteren Rollen als Leiter des Massenspektrometrielabors und IT-Koordinator des Instituts für Mikrobiologie und Virologie verbinde ich neu aufkommende wissenschaftliche Methoden wie beispielsweise die MALDI-TOF Massenspektrometrie v.a. mit meinem mykologischen Interesse. Anhand verschiedener bakterieller Genera (Campylobacter, Salmonella enterica, Clostridioides difficile) haben wir (Kooperation v.a. mit Prof. AE Zautner) zeigen können, dass sich anhand von MALDI-TOF-MS Daten auch unterhalb der Speziesgrenze noch verschiedene Subtypen differenzieren lassen die mit unterschiedlichen relevanten Phänotpyen korrelieren. Für verschiedene weitere pathogene Spezies generieren wir derzeit solche formalen Typisierungsschemata („mass spectrometric phyloproteomics, „MSPP“). In einem aktuellen Projekt (Kooperation mit Prof. AC Hauschild beschäftigen wir uns darüber hinaus mit der Entwicklung von KI-basierten Assistenzsystemen, um aus Massenspektrometrie-Daten durch Verknüpfung mit lokalen epidemiologischen Daten Resistenzphänotypen bei Bakterien vorherzusagen.
Ausgewählte Publikationen:
M Bernhard, N Worasilchai, M Kangogo, WL Trzaska, C Bii, M Weig, U Groß, A Chindamporn, and O Bader (Apr 2021). CryptoType – Public Datasets for MALDI-TOF-MS Based Differentiation of Cryptococcus neoformans/gattii Complexes. Frontiers in Cellular and Infection Biology 11:634382. doi: 10.3389/fcimb.2021.634382. [Pubmed]
C Noll, A Nasruddin-Yekta, P Sternisek, M Weig, U Groß, AF Schilling, FT Beil, and O Bader (Dec 2020). Rapid direct detection of pathogens for diagnosis of joint infections by MALDI-TOF MS after liquid enrichment in the BacT/Alert blood culture system. PLOS ONE 15(12):e0243790. doi: 10.1371/journal.pone.0243790. [Pubmed]
O Bader (Nov 2017). Fungal Species Identification by MALDI-ToF Mass Spectrometry. Methods Mol Biol 1508:323-337. doi: 10.1007/978-1-4939-6515-1_19. [Pubmed]
AE Zautner, WO Masanta, M Weig, U Groß, and O Bader (2015) Mass Spectrometry-based PhyloProteomics (MSPP): A novel microbial typing Method. Sci Rep 5:13431. doi: 10.1038/srep13431. [Pubmed]
Externe Kooperationen
Über unsere eigenen Fragestellungen hinaus haben wir immer wieder verschiedene Abteilungen der UMG und anderen Universitäten bei kleineren oder größeren diagnostisch-epidemiologischen Projekten unterstützt, und für verschiedene Industriepartner mikrobiologische Auftragsforschung durchgeführt.
Ausgewählte Publikationen:
S Taudien, W Leszczynski, T Mayer, U Loderstädt, O Bader, M Kaase, and S. Scheithauer (Mar 2023) Misidentification as Pseudomonas aeruginosa in hospital water supply samples. J Hosp Infect 133:23e27. doi: 10.1016/j.jhin.2022.12.013. [Pubmed]
M Kreutz, C Kreutz, P Kanzow, T Tauböck, P Dieckmann, C Noll, O Bader, B Rohland, A Wiegand, and M Rizk (Nov 2022) Effect of bioactive and antimicrobial nanoparticles on properties and applicability of dental adhesives. Nanomaterials 12(21):3862. Doi: 10.3390/nano12213862. [Pubmed].
S Dierks§, O Bader§, J Schwanbeck, U Groß, M Weig, K Mese, R Lugert, W Bohne, A Hahn, N Feltgen, S Torkieh, FR Denker, P Lauermann, MW Storch, H Frickmann, and AE Zautner (May 2021) Diagnosing SARS-CoV-2 with Antigen Testing, Transcription-Mediated Amplification and Real-Time PCR. Journal of Clinical Medicine 10(11), 2404. doi: 10.3390/jcm10112404 [Pubmed].
R Nißler, O Bader, M Dohmen, SG Walter, C Noll, G Selvaggio, U Groß, and S Kruss (Nov 2020). Remote near infrared detection and identification of pathogens with multiplexed nanosensors. Nature Commun 11(1): 5995. doi: 10.1038/s41467-020-19718-5. [Pubmed]
A Kauffels, G Schmalz, O Kollmar, J Slotta, M Weig, U Groß, O Bader, and D Ziebolz (2017). Oral findings and dental behavior before and after liver transplantation – a single center cross-sectional study. Int Dent J 67(4):244-251. doi: 10.1111/idj.12290 [Pubmed]
§: „equal contribution“
Mitglieder und Alumni der AG (~ab 2018)
Externe Kooperationspartner in aktuellen Projekten
aktuell
- Agnieszka Goretzki (ltd. MTA)
- Annekatrin Cordts
- Josefina Lovatto (DAAD)
- Marlene Jenderek (HiWi)
- Dr. rer. nat Pia Sternisek
- Marcel Schöne, MSc
- Dr. med. Claudia Folba
- Dr. med. Mareike Bernhard
- Carla Vogeler, BSc
- Dr. med Azadda Nasruddin-Yekta
- Nina Gerkens (Laborantin)
- Dr. rer. nat. Emilia Gomez-Molero
- Abdelmalek Salem Ahlees, BSc
- Yvonne Laukat (Laborantin)
- Felix Yoppe, MSc
Seite aktualisiert am: 21.03.2025
- Prof. Julian Naglik (King’s College, London, UK)
- Prof. Toni Gabaldon (BRC, Barcelona, ES)
- Prof. Piet deGroot (CRIB, Ablacete, ES)
- Dr. Jonathan Richardson (King’s College, London, UK)
- Jun. Prof Anne-Christine Hauschild (UMG, Göttingen, DE)
- Prof. Timo Buhl (UMG, Göttingen, DE)
- Dr. Christoph Sasse (GAUG, Göttingen, DE)
- jun. Prof. Lydia Herzel (FU Berlin)